| | ||||
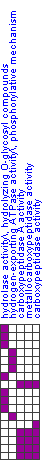 |
|
|||
 >4x >4x |
Module 492 |
| General Properties | |
|---|---|
| Below we list the number of gene sets from which the module was composed was originally composed, and the number of genes and experiments that it contains. Clicking on the number of genes will display a detailed list of all the genes in the module. Clicking on the number of experiments will display a detailed list of all the experiments in the module. | |
| Num gene sets: | 5 |
| Num genes: | 17 (see 479 additional genes for this module) |
| Num experiments: | 46 (29 induced, 17 repressed) |
| Parent module: | 568 |
| Children modules: | 402 |
| Gene sets | |
|---|---|
| Below we list the gene sets from which the module was originally composed. Clicking on a gene set will display all the genes associated with that gene set. Also listed are all the other modules in which the gene set participates. | |
| hydrogen_exporting ATPase activity, phosphorylative mechanism | |
| carboxypeptidase activity | |
| metallocarboxypeptidase activity | |
| carboxypeptidase A activity | |
| hydrolase activity, hydrolyzing O_glycosyl compounds | |
| Enriched clinical annotations | |||||||
|---|---|---|---|---|---|---|---|
| For each of the clinical annotations, we tested whether it was enriched in the set of arrays in which the module is significantly induced (or repressed). Below we list all such annotations that were enriched in this module with an FDR corrected p-value of 0.05 ('P-value' column), along with the number ('Hits' column) of induced (or repressed) module's arrays in which they appear. We also show the fraction of the module's induced (or repressed) arrays with the given annotation ('Hits (%)' column). For completeness, we also list the total number of relevant induced arrays in the module ('Module hits in category' column), total number of arrays in the compendium in which the corresponding annotation is present ('Arrays in annotation' column), and the total number of relevant arrays in the compendium ('Arrays' column). Note that the 'relevant' arrays may be different for each annotation, since it includes only arrays in which the annotation was relevant (i.e., present or not present) and excludes arrays where the value of the annotation was not known. Rows that correspond to annotations that were enriched in the induced arrays of the module have a red background, while rows corresponding to annotations enriched in the repressed arrays have a green background. For each annotation, we also list the other modules in which it was enriched. | |||||||
| Clinical annotation | Category | P-value | Hits | Hits(%) | Module hits in category | Arrays in annotation | Arrays |
| Melanoma cell line (NCI60) | NCI60 | 1.5e-10 | 10 | 90.9 | 11 | 16 | 139 |
| Melanoma tumor or cell line (NCI60) | NCI60 | 1.5e-10 | 10 | 90.9 | 11 | 16 | 139 |
| Hematologic cancer and cell line (Stimulated immune*) | Stimulated immune* | 2.7e-08 | 9 | 31.0 | 29 | 53 | 1945 |
| Hematologic samples and cell lines (Stimulated immune*) | Stimulated immune* | 2.7e-08 | 9 | 31.0 | 29 | 53 | 1945 |
| Macrophages (Stimulated immune*) | Stimulated immune* | 2.7e-08 | 9 | 31.0 | 29 | 53 | 1945 |
| Monocytes (Stimulated immune*) | Stimulated immune* | 2.7e-08 | 9 | 31.0 | 29 | 53 | 1945 |
| Cell line (NCI60*) | NCI60* | 1.4e-06 | 11 | 37.9 | 29 | 135 | 1945 |
| Cancer and cell line (NCI60*) | NCI60* | 1.7e-06 | 11 | 37.9 | 29 | 138 | 1945 |
| Hematologic cancer and cell line (Stimulated immune*) | Stimulated immune* | 5.9e-05 | 5 | 29.4 | 17 | 53 | 1945 |
| Hematologic samples and cell lines (Stimulated immune*) | Stimulated immune* | 5.9e-05 | 5 | 29.4 | 17 | 53 | 1945 |
| Macrophages (Stimulated immune*) | Stimulated immune* | 5.9e-05 | 5 | 29.4 | 17 | 53 | 1945 |
| Monocytes (Stimulated immune*) | Stimulated immune* | 5.9e-05 | 5 | 29.4 | 17 | 53 | 1945 |
| Enriched GO annotations | ||||||
|---|---|---|---|---|---|---|
| For each GO annotation, we tested whether it was enriched in the set of genes of the module. Below we list all such annotations that were enriched in this module with an FDR corrected p-value of 0.05 ('P-value' column), along with the number ('Hits' column) of the module's genes in which they appear. We also show the fraction of the module's genes with the given GO annotation that are included in this module ('Hits (%)' column). For completeness, we also list the total number of genes in the module ('Module genes' column), total number of genes in the compendium in which the corresponding annotation is present ('Genes in annotation' column), and the total number of genes in the compendium ('Genes' column). | ||||||
| GO annotation | P-value | Hits | Hits(%) | Module genes | Genes in annotation | Genes |
| hydrolase activity | 8.2e-14 | 17 | 100 | 17 | 783 | 4566 |
| carboxypeptidase activity | 1.6e-12 | 6 | 35.2 | 17 | 13 | 4566 |
| hydrolase activity\, hydrolyzing O-glycosyl compounds | 2.0e-12 | 7 | 41.1 | 17 | 27 | 4566 |
| regulation of metabolism | 7.8e-12 | 13 | 76.4 | 17 | 365 | 4566 |
| lysosome | 7.9e-12 | 8 | 47.0 | 17 | 57 | 4566 |
| hydrolase activity\, acting on glycosyl bonds | 4.1e-11 | 7 | 41.1 | 17 | 40 | 4566 |
| exopeptidase activity | 5.1e-11 | 6 | 35.2 | 17 | 21 | 4566 |
| carboxypeptidase A activity | 9.3e-11 | 5 | 29.4 | 17 | 10 | 4566 |
| metallocarboxypeptidase activity | 1.7e-10 | 5 | 29.4 | 17 | 11 | 4566 |
| hydrogen-exporting ATPase activity\, phosphorylative mechanism | 1.9e-09 | 4 | 23.5 | 17 | 6 | 4566 |
| metalloexopeptidase activity | 7.3e-09 | 5 | 29.4 | 17 | 21 | 4566 |
| vacuolar hydrogen-transporting ATPase | 2.7e-08 | 4 | 23.5 | 17 | 10 | 4566 |
| catalytic activity | 3.8e-08 | 17 | 100 | 17 | 1677 | 4566 |
| nucleoside monophosphate phosphorylation | 3.8e-07 | 4 | 23.5 | 17 | 18 | 4566 |
| hexosaminidase activity | 1.4e-06 | 3 | 17.6 | 17 | 7 | 4566 |
| membrane lipid catabolism | 2.3e-06 | 3 | 17.6 | 17 | 8 | 4566 |
| sphingolipid catabolism | 2.3e-06 | 3 | 17.6 | 17 | 8 | 4566 |
| carbohydrate metabolism | 3.2e-06 | 6 | 35.2 | 17 | 123 | 4566 |
| metallopeptidase activity | 3.9e-06 | 5 | 29.4 | 17 | 70 | 4566 |
| proton transport | 6.4e-06 | 4 | 23.5 | 17 | 35 | 4566 |
| regulation of catabolism | 1.6e-05 | 6 | 35.2 | 17 | 162 | 4566 |
| cation transporter activity | 1.7e-05 | 4 | 23.5 | 17 | 45 | 4566 |
| proteolysis and peptidolysis | 2.6e-05 | 6 | 35.2 | 17 | 176 | 4566 |
| hydrogen ion transporter activity | 2.7e-05 | 4 | 23.5 | 17 | 50 | 4566 |
| peptidase activity | 4.4e-05 | 6 | 35.2 | 17 | 193 | 4566 |
| sterol regulatory element-binding protease activity | 5.4e-05 | 3 | 17.6 | 17 | 21 | 4566 |
| nucleotide metabolism | 7.2e-05 | 4 | 23.5 | 17 | 64 | 4566 |
| catabolism | 8.1e-05 | 6 | 35.2 | 17 | 215 | 4566 |
| Visual display | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Below is a visual display of the module. Shown are those arrays in which the module's genes significantly change, and the direction of change (induction/repression) in each array is indicated (middle, labeled 'Module changes'; red/green row). The arrays that correspond to the significant clinical attributes are also shown (top; brown rows). The gene sets that compose the module are also shown (left) along with an indication of the membership of the module genes in these gene sets. | |||||||||
| You can also view the images within GeneXPress by loading the module gxp file file. | |||||||||
|
|||||||||