Region of Interest Based Medical Image Compression
Salih Burak Gokturk
Abstract
CT or MRI Medical imaging produces digital form of human body pictures. There exists a need for compression of these images for storage and communication purposes. Current compression schemes provide a very high compression rate with a considerable loss of quality. In medicine, it is necessary to have high image quality in region of interest, i.e. diagnostically important regions. This paper discusses a hybrid model of lossless compression in region of interest with high compression rate lossy compression in other regions. The hybrid technique provides efficient and accurate coding of the medical images. We first survey a number of lossless and lossy compression schemes for the hybrid coder. An application to abdomen CT imaging with the special interest on human colon is presented.
1. Introduction
Medical imaging has a great impact on diagnosis of diseases and preparation to surgery. On the other hand, the storage and transmission is an important dilemma due to enormous size of medical image data. For example, each slice of CT abdomen images is 512 by 512 of 16 bits, and the data set consists of 200 to 400 images leading to 150 MB of data in average. An efficient compression of the medical data can solve the storage and transmission problem.
Current compression schemes bring great compression rates if loss of quality is affordable. Medicine can not afford deficiency in diagnostically important regions ('Region of Interest'). An approach that brings high compression rate with good quality in region of interest (ROI) is required. A hybrid coding scheme seems to be the only solution to this twofold problem. In other words, two different compression schemes should be used for ROI and non-ROI. The general theme is to preserve quality in diagnostically critical regions, while encoding the other regions so that the viewer can observe the position of the critical regions in the original image. Therefore, a very lossy compression scheme is suitable in non-ROI regions to give a global picture to the user while a lossless compression scheme is necessary for ROI regions.
CT images can be considered as many 2-D images (slices) traveling in the human body. In some sense, they are images from a movie of very smooth but everywhere transitions. For high quality, lossless compression purposes, we exploit two types of redundancy: Spatial redundancy, i.e. in each slice of the data set and temporal redundancy, i.e. between consecutive slices in the data set. It has been shown that, the temporal redundancy reduction results with a slightly better entropy rate. We surveyed a number of lossy compression schemes for non-ROI. In section 2, we present the results of DCT based, principal component analysis based, blockwise vector quantization based, and motion coding compression schemes. In our experiments, motion compansated coding scheme resulted with the best compression rate for non-ROI regions.
The overall goal of this research, is to represent the images with the smallest possible number of bits and with no loss in ROI. For this purpose, a complete hybrid coder that uses motion compensated coder in the overall image and entropy minimizing lossless coder for coding the error in the ROI region is implemented. The experiments are applied on CT abdomen images with the special concern on human colon. The first step of a ROI based system is segmentation. In our application, the colon wall is segmented through a sequence of 3-D morphological image processing techniques.
Colon cancer is the second leading cause of cancer deaths in USA. To our knowledge, however, this is one of the first work concerning on compression of human colon CT data. The main research in this area is on graphical visualization of the colon and automatic colon cancer detection[12][13]. However, that research suffers from lack of data, mainly due to storage and communication problems. The development of compression technology would allow for efficient use of visualization and automatic detection techniques in human colon analysis.
The paper goes on as follows: In section 1.1, the previous work is explored. In section 2, a description of the lossy and lossless compression schemes is given, and the results for abdomen CT image compression are compared. Section 3 describes our proposed hybrid model solution and summarizes the results. Section 4 gives our conclusions and discusses the possible future work.
1.1. Previous Work
Since the evolution of digital medical imaging techniques, many researchers have attempted to apply compression methods. The initial concern was information preserving methods with the highest possible compression rate. Scan pixel difference was researched by Takaya et al in [1]. Assche et al exploits the interframe redundancies in [2]. Linear predictive coding schemes were investigated in [3]. One of the main observations in all of these studies was the low compression rate.
In terms of transform coding, Principal component analysis was given as a better alternative to DCT based compression schemes in [8] and [9]. The main assumption for this is the statistical similarity that the medical images would exhibit. However, we achieved better compression rates with DCT compared to principal component analysis in our experiments.
In order to achieve hgher compression rates without paying off from quality, region of interest based methods were investigated in the following years. In [4], a ROI-DCT algorithm that uses more DCT coefficients in ROI, was proposed. A subband compression scheme was used by Cosman et al. in [5]and [6]. In [7], 3-D wavelet compression is investigated. The most important drawback of 3-D based approaches in ROI based compression is twofold: First, the resolution between the slices is much less than the resolution in the image plane. Second, the ROI does not necessarily lie in a 3-D primitive shape such as a cube. Due to these reasons, 2-D ROI based scheme is explored in this paper.
2. A comparison study of compression schemes applied to CT abdomen images
In this section, we describe and evaluate the following compression schemes: 1- Lossless Compression Schemes, 2- DCT based compression scheme 3- Principal Component Analysis based Compression Schemes 4- Blockwise Vector Quantization 5- Motion compansated compression schemes. The optimal of the above methods, i.e. motion compensated coding, is proposed in a ROI based hybrid compression scheme in section 3. It has to be noted that, for all of the CT images used in experiments, the uncompressed data rate is 16 bpp.
2.1. Entropy minimizing Lossless Coding
The entropy is referred as the minimal number of bits to represent the information. The formulation is given as follows:
![]()
where pk is the probability of a particular intensity. The variable length coding schemes such as Huffman Coding or arithmetic coding approximates the theoretical entropy. The theoretical entropy of the intensity values of the CT abdomen images is found as 7.93 bits. However, there is statistical dependency between the values of neighbor pixels in the images and should be exploited.
An example of two consecutive CT abdomen images and their difference is given in figure 1. To exploit the redundancies in inter-slices, a coding scheme that predicts the current pixel as a linear combination of the west, north and north-west pixels is used. The prediction function and the error function is given as follows:
![]()
![]()
where Ip is the predicted image and e is the error image. The entropy of the error vanishes to 5.9 bits. To achieve bigger compression rates, the temporal statistical dependency of the pixel values is considered. In other words, each image pixel value is predicted as the same pixel value in the previous image such as:
![]()
where the superscript t denotes the slice number (time). The predictive error entropy reduces to 5.76 bits. This result also suggests that there is more temporal redundancy than the spatial redundancy in CT abdomen images. However, when we look at the difference images, we observe that more of the information is included in the ROI area.

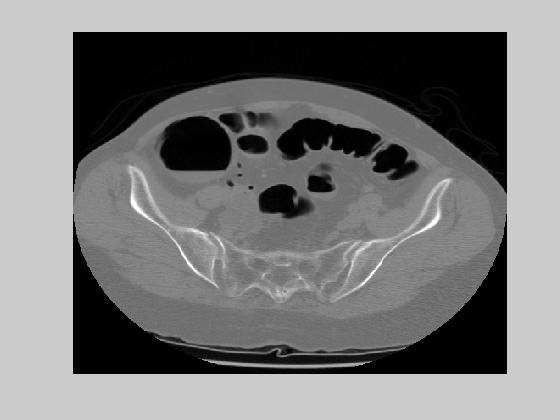
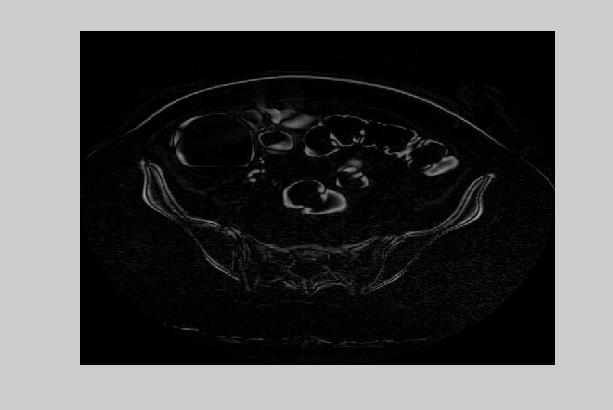
Figure 1. Two consecutive CT abdomen images and their difference image
2.2. DCT based Compression
Discrete Cosine Transformation (DCT) is a powerful mathematical tool that took its place in many compression standarts such as JPEG and MPEG. In the most general form, DCT can be expressed as matrix multiplication. The mathematical definition of 8 by 8 DCT matrix is given as follows:
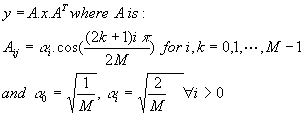
where M is the block-edge size, x is M by M input image matrix, and y is M by M DCT coefficients.
We evaluated blockwise 8 by 8 DCT on CT abdomen images. The block diagram of DCT processing is given in figure 2. In the experiments, the theoretical entropy values of 64 DCT coefficients are calculated. We used two different scheme for coding the DCT coefficients. First involves entropy minimizing coding of the DCT coeeficients. Second uses Run Level Coding of the DCT coeeficients while still using entropy minimizing coding. It has been shown that the use of Run Level Coding with Entropy minimizing coding increases the compression rate for highly lossy DCT compression. The mean squared error values are calculated for different quantization levels. Figure 3, summarizes the results for two cases: Using only entropy minimizing coder vs Using run level coder with entropy minimizer coder.

Figure 2. DCT Transform Encoder and Decoder Block Diagram
| Quantization
Step Size |
1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 |
| MSE in dB | -11.7 | -5.7 | 0.34 | 6.26 | 11.93 | 17.18 | 21.81 | 25.73 | 29.31 | 32.64 | 35.95 |
| Rate (without RLC) (bpp) | 5.74 | 4.97 | 4.09 | 3.20 | 2.34 | 1.57 | 0.96 | 0.55 | 0.31 | 0.16 | 0.09 |
| Rate (with RLC) (bpp) | 8.04 | 7.09 | 5.87 | 4.51 | 3.15 | 1.95 | 1.07 | 0.55 | 0.28 | 0.14 | 0.07 |
Figure 3. The Mean Square Error vs Bit Rate: with or without RLC
DCT is a very promising technique that can be used either on the non-ROI for a very big compression rate, or on ROI for higher quality. Figure 4 gives examples of reconstructed images that is mapped to 256 gray scale levels. The main drawback of DCT transformation is its time complexity. The decoder has to involve many matrix operations to reconstruct the image. In movie or medical image compression, the simplicity of the decoder is very important. Similar to MPEG standart, DCT is more likely to be used as a secondary engine in medical image compression.

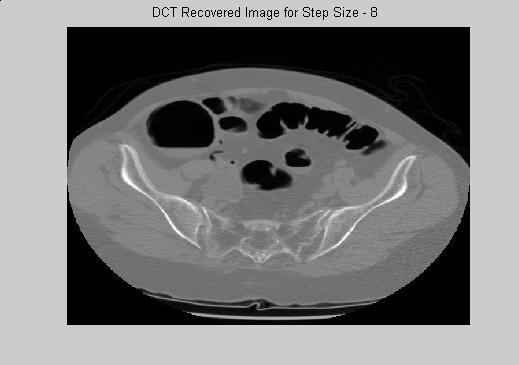

(a) (b) (c)

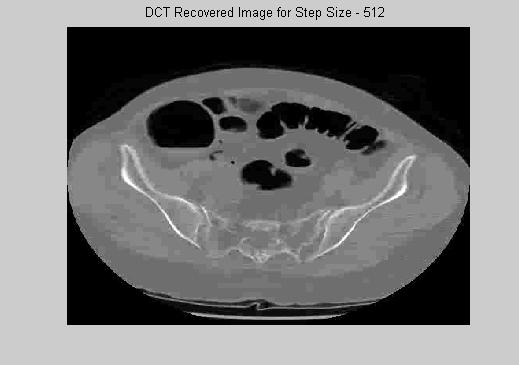

(d) (e) (f)
Figure 4. DCT Reconstructed images for stepsize (a) to (f) : 2, 8, 32, 128, 512, 1024
2.3. Principal Component Analysis (PCA) based Compression
Principal component analysis is a method for estimating optimally a low dimensional representation of data embedded in a high dimensional space. In other words, PCA extracts the most important features of a high dimensional space in the sense that the projection error onto those feature spaces is minimal. PCA takes its place in many applications such as detection, recognition, image processing and compression [10]. In mathematical terms, Principal components are the eigen vectors of the autocorrelation matrix of the data. We have obtained a 256-vector from each 16 by 16 image block by stacking the column pixels in the block. Let Xk be the column vector obtained for each block k. The principal components of the set of all block vectors is obtained as follows:
![]()
![]()
where R is the total number of blocks, X0 is the average vector and X'k is the average subtracted vector. Next, we form the matrix A, columns of which are X'k and apply SVD on A:
![]()
![]()
where U and V are 256 by 256 and R by R ortonormal matrices. The principal modes are given by the columns of the matrix U. To reduce the rank of the matrix A in an optimum manner such that most of the energy still resides in the data, we can represent the data by the first N columns of U. Let uk be the kth column of the matrix U. The low rank representation can be expressed as follows:
![]()
where ßi are the weights of principal components. In other words, a block image can be represented by a number (N) of the principal components where the projection error (Xp-X) is minimized in the least square sense. This way, each image block is compressed by being represented by only a few parameters ßi. Figure 5 shows the reconstructed image that uses 16 by 16 block size and N=31 first principal components. Although the images look as pleasing as DCT images, the MSE error for N=31 was found as ~30dB which is higher compared to DCT. This result proves the following: PCA minimizes the error in the linear, least square manner, however, the CT abdomen block images do span a nonlinear space, and DCT extracts the nonlinearity of this space better than PCA.
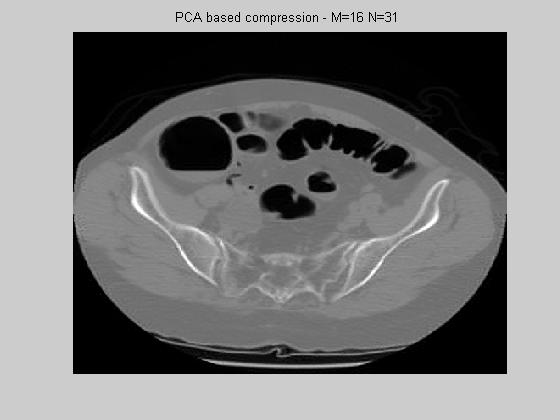
Figure 5. PCA using 16 by 16 blocks and the first 31 principal components
2.4. Blockwise Vector Quantization
Medical 3D volume images contain many 2-D slices. Therefore, the decoder side of the compression setup should be simple. DCT and PCA compression involve many multiplications in the decoder side. Besides that, the non-ROI can be compressed in a very lossy way but keep the global view of the slice. In this and the following section, two methods that have very simple decoder design are presented.
A straightforward approach is coding only the average of the blocks in the image. The decoder linearly interpolates between the block centers and reconstructs the image. In other words, the reconstructed images are formed by convolving the sampled and lowpass filtered image by a pyramid. The flow of the processing is given in figure 6.

Figure 6. Flow of blockwise vector quantization
Once the average is extracted for each block, Lloyd Max quantizer is executed. In our experiments, the centroid of each quantization level was found by the K-means algorithm with K=32. The theoretical entropy of these 32 levels of quantization was found to be 3.69. We experimented the setup with M=5 and M=7. The corresponding rates are 0.14 and 0.075 for M=5 and M=7 respectively. The reconstructed images for each case are presented in figure 7. The mean squared error for these images are 38 dB and 39 dB. Although the mean squared errors are much worse compared to DCT, the global view is still comprehensible. The main advantage of this approach is the simplicity of the decoder, since the decoder is composed only of a linear interpolation engine (at most four multiplications per pixel.).
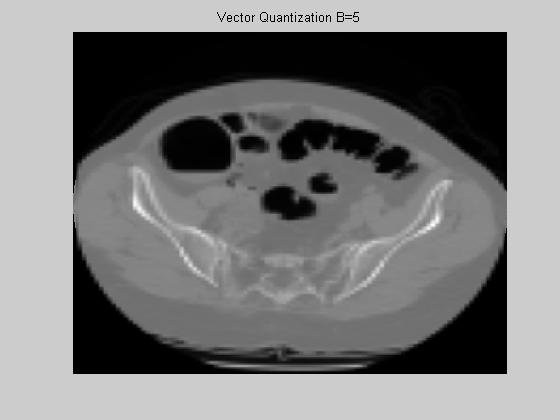

(a) (b)
Figure 7. Vector Quantization - Reconstructed image with block size: (a) 5x5 (b) 7x7
2.5. Motion Compensated Hybrid Coding
The main theme behind motion compensated coding is the smooth movement of objects between two consecutive slices. First, each slice is partitioned into blocks. Next, motion vector for each block is found using the previous image. In other words, for each block in the current image, a same size block is predicted from the previous image. However, if the error between the predicted block and the current block is big, another compression scheme, i.e. lossless compression, is used for the prediction of the current block. The flow diagram of the motion compensated hybrid encoder is given in figure 8. The decoder is very simple and is composed of either block shifting from the previous image or lossless decoding.
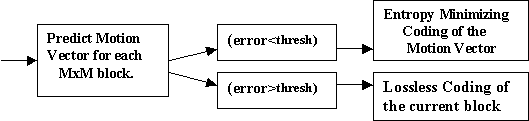
Figure 8. The diagram for the motion compensated hybrid encoder.
Motion prediction is one of the important steps in motion compensated coding. For translational motion prediction, motion vector is composed of two components: translation in x (dx) and translation in y(dy) . In this research, we have chosen to use Lukas-Tomasi-Kanade optical flow tracker[11]. The goal of the Lukas-Kanade tracker is to minimize the difference of current image block with the translated image block. The to-be-minimized energy function can be expressed as follows:
![]()
where I1 and I2 represent the two consecutive slices. The solution to this problem is found by taking the derivative of the energy function (E) with respect to dx and dy :
![]()
The first order linear approximation to I2(x+dx,y+dy) is given as follows:
![]()
Putting this expression in the equation for derivative of E, and setting it to 0:
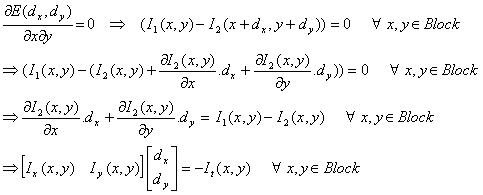
Where Ix and Iy are spatial and It is temporal derivative of the images. Observe that the final equation relies totally on the first order linear approximation for I2(x+dx,y+dy). This approximation holds very well for small values of dx and dy. However when dx and dy become more than 1 pixel, the approximation does not hold well in its original form. In order to be able to approximate for higher values of motion vector, the main equation is iterated in a gradient descent manner until the change in dx and dy become less than a threshold. In our experiments, we used a threshold of 0.1 pixels, that is to say, the estimation of dx and dy are in a 0.1 pixel accuracy.
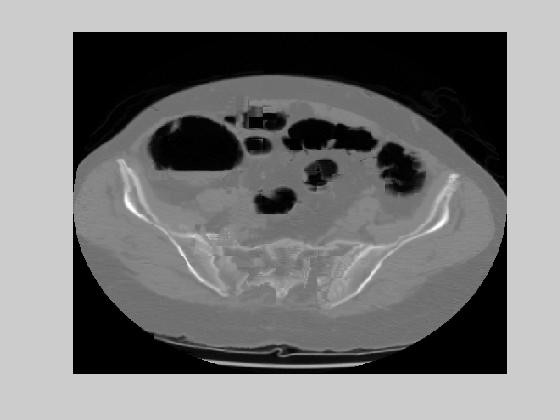
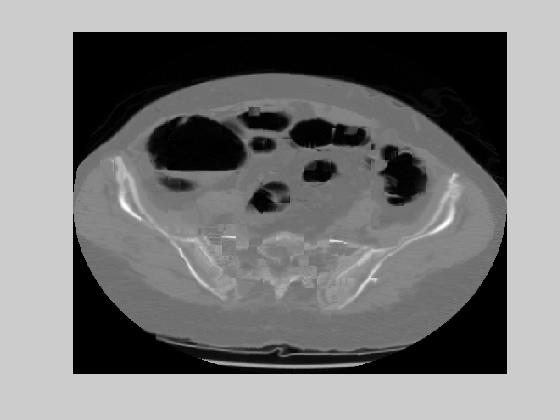
We used a block size of 16 by 16. If the rms error for a block becomes more than 40 dB, that block is coded with an entropy minimizing lossless encoder. Figure 9(a) shows the motion vectors for the CT image of Figure 1. Figure 9(b) and 9(c) show two examples of the reconstructed CT slices with the original images given at Figure 9(d) and 9(e). The performance is negatively effected on the edges of the image. For an accuracy of 0.1 pixels, the theoretical entropy of the motion vectors was found as 2.28 and 2.45 in dx and dy respectively. This result brings a bit rate of 0.018 bpp. The average rms error was 35 dB. These results show that medical images are very appropriate for motion compensated coding. The main reason behind this is the smooth transition of images throughout the CT slices. Next section will discuss the application of motion compensated coding in a region of interest based compression system.

(a) (b) (c)


(d) (e)
Figure 9. (a) Motion field example (b)-(c) Reconstructed images for the originals given at (d)-(e).
3. Region of Interest Based Compression of Medical Images
This section describes a hybrid compression system for lossless compression of region of interest in CT abdomen images. The colon wall is chosen as the region of interest. The whole image is first coded by motion estimating compression. This acts also as an initial guess for ROI areas. For ROI, the difference of the initial guess and the original image is coded by an entropy minimizing lossless compressor. The details of the system is described in section 3.2. First we describe the segmentation of colon from CT abdomen images in section 3.1.
3.1. Segmentation using morphology
Mathematical morphology is a branch of science which is built upon set theory and has many application areas in image processing. This approach includes generation of mappings for each pixel according to the pixel's local neighborhood. Many researchers have used this technique to segment biomedical images[14][15]. Segmenting the colon from the CT data set is composed of three steps:
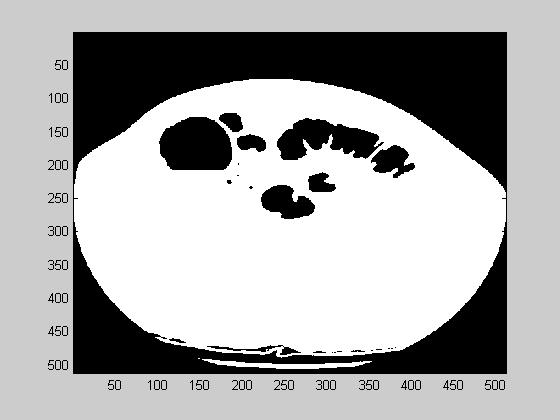
- First, the air is segmented from the tissue. This is a thresholding operation, and the result of this step is shown at image 10(a). Air is shown in black in this image.
- Next, the colon wall that surrounds the air is extracted. A derivative operation is suitable for this task. We used a 3D version of Sobel derivative operator. The gradient magnitude is shown in figure 10(b). The derivative is shown in white.
- The region of interest is given as 5 pixels inside and outside of the colon wall. A morphological 3D grassfire operation is suitable. This algorithm, finds points that are equal distance from a layer of points. In other words it is composed of a series of dilation operations. The final segmented image, that contains 5 pixels of ROI around colon wall is given in figure 10(c). The image brightness is max at the colon wall and decreases as it departs from the colon wall.
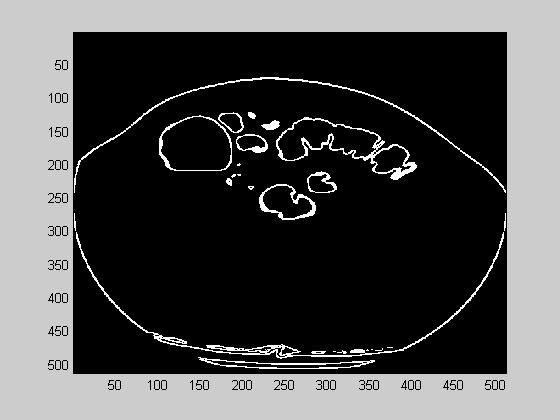

(a) (b) (c)
Figure 10. (a). Air segmented image (b). The derivative of (a) (c) Final segmented image
3.2. Region of Interest(ROI) Based Compression System
Once the ROI is segmented from each slice, a hybrid compression scheme is used for coding each slice. The flow diagram of the encoder is given in figure 11. First, the image is compressed by motion compensated coding. We also assume that the initial slice of the volume is compressed in a lossless manner. Next the following algorithm is applied for each block:
- The mse between the original image block and the motion estimated block is found.
- if mse is lower than high threshold and the block does not contain any ROI, or mse is lower than the low threshold and the block contains ROI than only the motion vectors are used for that block.
- else if block contains ROI and mse is higher than the low-threshold or block does not contain ROI but mse is higher than the high-threshold, additional entropy minimized lossless coding of the block is required. We have chosen to take the motion estimated block as the initial guess, and coded only the difference between the original block and the motion estimated block. This way, the theoretical entropy of the difference blocks was calculated as 4.38, which is better than the value obtained in section 2.1.

Figure 11. Block diagram of the ROI based compressor
The experimental values of low and high threshold were 20 dB and 40 dB respectively. The setup was tested by blocksize of 16 by 16 and 8 by 8. The bigger the block size, the better is the motion compensated coder rate. However, the bigger the blocksize, the bigger is the ratio of ROI blocks. Experiment with 16 by 16 blocksize produced %12.2 ROI blocks. The theoretical entropy of motion vectors was 2.28 and 2.45 bits in x and y directions respectively.. This introduces a bit rate of 0.018. The theoretical entropy of the error images turned out to be 4.38bits. The average rate of the ROI based hybrid coder becomes %12.2*4.38+0.018=0.552bpp. This came with an average rms error of 33.7 with lossless coding in ROI. Reconstructed images that correspond to images on figure 9(b) and (c) are given in figure 12(a) and (b). Experiment with 8 by 8 blocksize produced %7.3 ROI blocks. The theoretical entropy of the motion vector was 1.82 and 1.96 in x and y directions respectively which brings a bit-rate of 0.059. The theoretical entropy of the error images was 4.31bits. The average rate of ROI based compression scheme with 8 by 8 blocks then become 0.3736. Considering that each pixel of CT slices is currently coded by 16 bits, the rate of compression is %2.3. The average rms error was 30.3dB with lossless in ROI. Figure 13(a) and 13(b) give the reconstructed images for the same slices with the use of 8 by 8 blocks. Both of the experiments were applied on a sequence of 20 slices. The reconstructed and original images can be seen here.


(a) (b)
Figure 12. ROI based compression results on the images of figure 9(b) and (c) with block size 16x16.
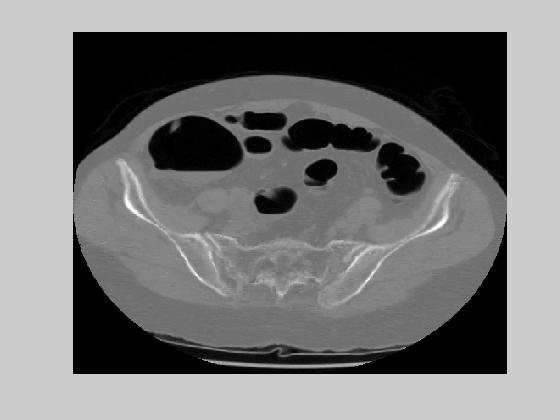
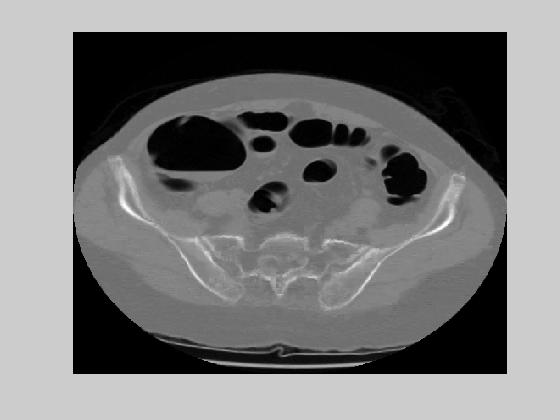
(a) (b)
Figure 13. ROI based compression results on the images of figure 9(b) and (c) with block size 8x8.
Observe that choice of 8x8 for block-size produced both high quality and better rate. The main reason for getting a better rate was the reduction on the proportion of ROI blocks with 8 by 8 blocks. If this proportion hadn't changed, 8x8 blocks would yield same quality but bigger rate images. The experiments show that a ROI based compression scheme produces a better quality and rate compared to the other methods that were explored in section 2.
4. Conclusion
In this study, we present a hybrid scheme that is appropriate for efficient and accurate compression of 3D medical images. The model uses lossless compression in region of interest, and very high rate compression in the other areas. After surveying through common compression schemes, we have chosen motion estimated coding as a prediction scheme for each medical slice. The difference of the ROI blocks and the prediction is coded separately with an entropy minimizing coder. We applied our experiments on CT abdomen images with the colon wall as ROI. The results show that a compression rate of %2.3 can be obtained by the ROI based compressor.
There are many possible directions of future investigations. We would like to evaluate the performance of affine tracker in stead of translational tracker. In order to get better compression rate, ROI can be lossy encoded. Future study will include a case study with the radiologists to observe the effect of lossy compression in ROI on diagnosis performance of the user.
Acknowledgements
We would like to acknowledge Bernd Girod, Markus Flierl and Sung-Won Yoon for the information given in class EE 368-B and also the directions that they gave throughout the research.
Bibliography & References:
1 Takaya,
K, Tannous, C.G., Information preserved guided
scan pixel difference coding for medical images,WESCANEX
95. Communications, Power, and Computing. Conference Proceedings., IEEE Volume:
1 , 1995 , Page(s): 238 -243 vol.1.
2
Van Assche, S.; De Rycke, D.; Philips,
W.; Lemahieu, I., Exploiting interframe redundancies in the lossless compression
of 3D medical images.Data Compression Conference, 2000. Proceedings. DCC 2000 ,
2000 , Page(s): 575.
3 Jian-Hong
Hu; Yao Wang; Cahill, P.T, Multispectral code excited linear prediction coding
and its application in magnetic resonance images, Image Processing, IEEE
Transactions on Volume: 6 11 , Nov. 1997 , Page(s): 1555 -1566.
4
A.Vlaciu, S. Lungu, N. Crisan, and S.Persa. New compression techniques for
storage and transmission of 2-D and 3-D medical images. In Advanced Image and
Video Communications and Storage Technologies, volume 2451, pages 370-7,
Amsterdam, Netherlands, March 1995..
5 Gray,
R.M.; Olshen, R.A.; Ikeda, D.; Cosman, P.C.; Perlmutter, S.; Nash, C.;
Perlmutter, K. Evaluating quality and utility in digital mammography, Image
Processing, 1995. Proceedings., International Conference on Volume: 2 , 1995 ,
Page(s): 5 -8 vol.2 .
6 Cosman,
P.C.; Gray, R.M.; Olshen, R.A. Evaluating quality of compressed medical images:
SNR, subjective rating, and diagnostic accuracy. Proceedings of the IEEE Volume:
82 6 , June 1994 , Page(s): 919 -932.
7 Baskurt, A.;
Peyrin, F.; Benoit-Cattin, H.; Goutte, R., Coding of medical images using 3D
wavelet decompositions, Acoustics, Speech, and Signal Processing, 1993.
ICASSP-93., 1993 IEEE International Conference on Volume: 5 , 1993 , Page(s):
562 -565 vol.5 .
8 Taur,
J.S.; Tao, C.W. Medical Image Compression using Principal Component Anlyasis,
Image Processing, 1996. Proceedings., International Conference on Volume: 1 ,
1996 , Page(s): 903 -906 vol.2.
9
Yoshioka, M.; Omatu, S. Image Compression by
nonlinear principal component analysis, Emerging Technologies and Factory
Automation, 1996. EFTA '96. Proceedings., 1996 IEEE Conference on Volume: 2 ,
1996 , Page(s): 704 -706 vol.2.
10
S.B. Gokturk J. Bouguet, R. Grzeszczuk, A Complete.System for Face
Tracking, Submitted to International Conference on Computer Vision (ICCV) 2001.
11 J.Shi,
C. Tomasi, Good features to track, International conference on computer vision
and pattern recognition, CVPR 1994.
12 S.B.
Gokturk, C. Tomasi, A graph method for the conservative detection of the polyps
in the colon. International Symposium on virtual colonoscopy., Boston, October
2000.
13
S.B.Gokturk, B.Acar, D. Paik, C. Tomasi, C. Beaulieu, S. Napel,
Recognising Polyps from 3D CT colon data, Biomedical Computation at Stanford
University, 2000.
14
R.Vogt, Precise Extration of Bones from CT Scans, Advances in
Mathematical Morphology, Volume 2.
15
J.Serra, Image Analysis and Mathematical Morphology, Academic Press,
New York, N.Y., 1982.