
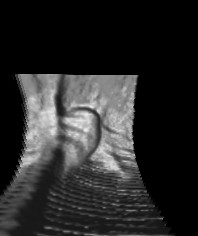
A survey on 3-D Segmentation, Modeling and Object Recognition
Salih Burak Gokturk
In this survey, I have focused mainly on the segmentation methods that can be applied to 3-D data processing namely 3-D medical volumes obtained by Computer Tomography (CT). The survey goes on with a description of the appropriate methods to model the 3-D shapes and how to use them for recognition of 3-D patterns in a set of candidate polyps. Document also include the conclusions regarding the appropriate selection of segmentation and modelling methods for processing of 3-D CT volumes.
Segmentation is extracting the information of a particular object from an image. This paper makes a summary of the segmentation methods that have been applied to 2-D and 3-D images. The segmentation methods can be classified in different ways on the basis of approaches adopted and the area of the image that the approach works on. The discussion aims to introduce the recent segmentation methods used in biomedical image processing and will purpose to explore the methods that can be applicable to segmentation of polyps from Computer Tomography Colon Images.
1. Introduction
Image segmentation is a branch of science that interested people since the discovery of cameras and image generation techniques. Many computer vision applications include processing of typical objects in a scene. Segmentation Results are aimed to give a global and/or local understanding of objects in the organization of a scene. Hence, image segmentation is a crucial step in the processing of images.
Many methods have been proposed for segmentation of images. The methods can be grouped in different ways according to the perspective of the view. One way of grouping the methods is according to the region of interest: 1- Boundary Based Methods 2- Region Based Methods[1]. A summary of typical applications will be given in the preceding sections.
Other ways of grouping the methods can be summarized as being according to the locality of the method, such as local vs. global methods[4], according to the frame in which we make the observations such as object vs. viewer centered approximations [3], or according to some other broad categories such as Low-level vs. High-level segmentation[2]. The preceding sections will discuss the applicability of these methods to volume based data structures.
The paper will mainly focus on the deformable models for the segmentation process. The use of energy minimizing curves namely snakes, to extract features of interest in images has first been introduced in [5]. Using a hybrid of local and global views, deformable models have been shown to converge to the expected structure in a few number of iterations[6][7][8].
The use of Fourier transformation and elliptic Fourier Descriptors have been shown to represent smooth structures with only a few coefficients[9]. The formulation of these techniques will be explained in detail in case of contours and surfaces.
The paper goes on as follows: In the second section, the document will summarize current favorite 2-D segmentation, modeling and matching methods and will discuss their applicability to 3-D applications. The third section will give a classification of the segmentation methods. The fourth section gives a brief formulation of deformable models including the snake and the balloon model. In the next section we will focus on the representation methods, which includes the polynomial based and orthogonal domain based methods. This section also gives information on the learning and matching algorithms that are applicable to these representation methods. Finally, we give our conclusions about the applicability of the explored methods to 3-D CT images (Figure 1).


Figure 1
2. Methods for 2-D Images
The 3-D data processing including the video and volume processing is a relatively new topic. Hence we will first give a description of the image segmentation, representation and matching methods in 2-D (ordinary) images.
Many methods have been proposed for the segmentation, modeling and recognition processes in 2-D. To increase the performance of these methods, initial simplification methods may be applied to images.[10][11] Template based methods have been used for appearance based identification of images. Methods such as region growing, contour following, connected component analysis can be used to identify the regions of an image[10][11]. Fourier Transform based image identification is attractive since high amount of information content in many images is in the lower band of frequency spectrum[12]. Line and shape descriptors will also be reviewed in this section.
2.1. Initial Simplification Methods
These methods are applied in order to simplify the images for further processing. This simplification can be in means of differentiation, smoothing or both. If the edges have the information content for further processing, differentiation based simplification can be applied. If the image seem to be noisy, the smoothing filters can be used to get rid of the noise. These can be implemented using convolution masks which consist of 2-D windows. Simplification methods can be applied to 3-D data by using 3-D convolution masks which represent a volume in space.
2.2. Template Matching
In many of the scene analysis problems, we would like to know if the scene includes a particular object. The classical technique employed to answer this question is 'Template Matching'. If g(i,j) is our image and t(i,j) is the template, the measure of how well a portion of an image matrices the template can be formulized as follows[10]:
![]()
Two other metrics that can be used are Euclidian based (L1 norm based) or the metric based on Linfinity norm. Statistical interpretations of this method have also been employed using priori probabilities of the objects in a scene. The correlation function based approach can also be applied for template matching. The core template matching approach does not take into account the effects of scaling and rotation of the object in an image, and different templates have to be applied for the detection of the rotation and/or scaling, which brings a bigger time complexity to the method.
2.3. Region Analysis Methods
Region based methods aim to distinguish and label different objects with respect to their regional intensity behaviors. These methods may be chosen in the cases where the intensity levels in the images have the information content for the segmentation process. Some of the region based methods can be summarized as follows :
2.4. Fourier Transform Analysis
Fourier transform is one of the tools to see the frequency changes in a signal. Since the convolution in spatial domain corresponds to the multiplication in frequency domain, Fourier Transformation has also been a perfect tool for mathematical analysis and formulations. The equation of the Fourier transform and its inverse transform in 1-D is given as follows[14]:
![]()
![]()
The formulation can easily be extended to 2-D and higher degrees. The Fourier transform of a signal has some typical properties regarding the translation, scaling and rotation of the signal. In 2-D, a simple translation of an image corresponds to a phase change in the Fourier domain, that is to say the magnitude of the Fourier Transform does not change. The scaling of an image corresponds to a scaling and a multiplication by a constant in frequency domain. The rotation of an image leads to the same amount of rotation in frequency domain. Another advantage of the Fourier Transform is that the convolutions in spatial domain correspond to multiplication in frequency domain namely filtering. Since the smooth images such as the medical images do not have many high frequency components, Frequency domain is a good choice of representing these images.
Fourier Transformation is also the major tool used in the theory of stochastic signal analysis, mean square estimation and Wiener Filtering[15]. For knowledge based applications where transmitted signal is estimated from its noisy version, Fourier Transformation is used as a tool for the mathematical formulation.
2.5. Shape Descriptions
In this section, different ways of line and shape description will be explored. The problem that is investigated is to come up with the best choice of shape to represent a number of points in an image. Below, a few ways to solution of line fitting problem are listed:
Many shape descriptors have been defined for modeling objects of interest. Below a very brief summary of most popular of them are given[10]:
![]()
The moment invariants, which are generated from the moment descriptors have been shown to be invariant to transformations such as rotation and translation on the image[16].
3. Classification of Segmentation Methods
Segmentation is grouping the image into parts where each part is homogenous with respect to one or more characteristics. In this section a brief classification and description of 3-D segmentation methods will be given. Many methods have been proposed for image segmentation. These methods have been grouped in different ways according to different measures. Some of these groupings are as follows:
Boundary based methods are built on the detection of local edges followed by an interpretation of the edges. Some examples of edge based applications will be explored in this section. Region Based methods, as described in section 2.3, map individual pixels of an image to pixel sets called regions. Neural Network based applications and graph theoretic approaches can be included in this group.
Local methods operate on the local parts of an image without using any information about the global content of the image. Convolution masks, or local edge detectors can be included in this group of methods. Global methods usually focus on features and objects in the image, and also concentrate on the whole image in stead of a local subpart of the image. Since the deformable models take the advantage of both of the approaches, it can be included in the hybrid methods.
The Low level segmentation includes pixel based, localized techniques such as thresholding, or Split and Merge Techniques. Medium level segmentation uses the scene domain cues besides the picture domain cues. These methods implement suitable representations for the scene domain cues such as spheres for polyp structures in a colon, and apply the segmentation according to these representations. Methods such as deformable model based segmentation, or graph theory based image segmentation can be included in this class. High level based segmentation methods include expert system-based segmentation.
Object centered representations attach a reference frame to an object and express the object geometry in that frame. The deformable representation model of snakes can be included in this class. Viewer centered representations model 3-D objects through a set of views taken in all possible conditions and transformations. Eigen spaces are an example of viewer centered representations.
Feature based techniques represent the objects through their features (edges, corners, area, volume, color). These methods generate compact object descriptors and they are more robust to transformations of an object (rotation, scaling, translation). The disadvantage is that they can not be applied to images and the features of the image should be extracted first. The image based applications work directly on the images.
This section will go on with a brief description of some segmentation methods. The deformable models will not be explored in this section but will be occupying the next section.
3.1. Eigen space Based Segmentation
This method can be used to find out the existence of an object in an image. Template based or correlation based image matching is not a time efficient and practical method if all views of an object can occur in an image. If we have a database of objects and their images from different views and in different conditions, we can simply form an eigen space based representation[3]. If x is the N3x1 vector representing an NxNxN volume, and if we have m views of the image x1, x2,...,xm, then we can formulize the set of vectors as follows:

where e1, e2,...,em are eigen vectors of the covariance matrix, Q=XXT and gji are the components of xj in the direction of ei. Using this approach and accounting for only the bigger eigen values, we can represent images with a vector g. After the database of the model images are formed, a new image is tested by comparing its g vector with the g vectors of library images. The closest distance matching g vector gives the output of the algorithm. In [17], Campbell and Flynn have proposed a description of eigen shapes for 3D object recognition in 3D where they extended the idea of eigen based recognition.
3.2. Fourier Domain Based Segmentation
Fourier domain has been extensively used for the representation of images. In [18], the formulation of 3-D fourier tranformation has been done and using the properties of transform, frequency domain representation of the the surface of a 3-D object is derived. The surface of the 3-D object V(x,y,z) can be derived by its gradient vector field:
![]()
Where kx, ky, kz are the unit vectors along x, y and z directions. The 3D DFT of the surface gradient is given by the frequency domain vector as follows:
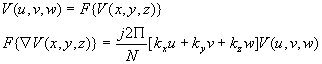
Using these formulas the frequency domain representation of the surfaces can be found without finding the surface gradient itself. The frequency domain representation can further be used for segmentation and matching purposes.
3.3. Morphology Based Segmentation
Mathematical morphology is a branch of science which is built upon set theory and has many application areas in image processing. This approach includes generation of mappings for each pixel according to the pixel's local neighborhood. Many researchers have used this technique to segment biomedical images. In [21], the 3D problem has been broken down to 2D problems, and mathematical morphology has been used for the segmentation of each slice.
A brief description of the mathematical morphology operations will be given in this section. The possible application areas will also be noted.
![]()
where X denotes the input set, B denotes the morphology window.
![]()
where C denotes the complement of the set.
![]()
![]()
These basic definitions of morphological operations can be extended to gray scale images [22] and high dimensional images [2]. The morphological operations can be used in the task of segmentation. For this, a search space is defined first. Using the appropriate morphological operations, the search space is constrained into a smaller subset where the surface elements are located in. Projection information from the 3D scan may also be used in this step. Next surface candidate elements are found and linked. For this step, the use of morphological scale space edge detection can be used. The final remaining step for surface segmentation is the reconstruction of the surface which is required for graphical interface.
3.4. Local Information Based Segmentation
Depending on the application, local information on the image can be used for segmentation purposes. This kind of segmentation usually uses specific information about the shapes, gradients, or intensity levels of the image. In [19], the differentiating intensity level of Corpus Callosum was use to segment it in the brain.
In medical images, segmentation using intensity values is limited due to field inhomogeneities. Edge based segmentation methods suffer from spurious edges and gaps in boundaries. In [23], a method which uses the edge based and region based segmentation is presented. Three dimensional MR brain segmentation was applied using explotion of local contrast followed by supervised segmentation technique by a minimal user interaction.
3D segmentation of Bone from CT images has been applied in [24]. The technique involved a tensor descriptor for each neighborhood, i.e. for each voxel in the data set. The shape of the tensors describe local structure of the neighborhood in terms of how much it is like a plane, a line, a sphere. This approach has been applied to segment the bone structure.
3.5. Model Guided Segmentation
Model guided segmentation approach uses the library models to segment the training set of images. Lundervold, Duta, Torfin and Jain proposed a new algorithm that uses both the prior information about shape and multispectral MRI measurements (intensity values) to segment Corpus Collasum with very little user interaction. Their shape learning stage used prior information about the mean shape of the corpus Callosum and its principal modes of variation.
This approach can be extended to any problem where the segmented training objects models are to be matched by the library models. Using the information from the database itself is a promising and efficient approach to the modeling and matching problem.
4. Deformable Models
Deformable models is a powerful tool in the segmentation of biomedical images[2]. Medical Images and volumes usually contain complex and irregular structures, hence segmentation and representation of these shapes with local descriptors is difficult. Deformable Models, namely snakes and balloons seem to be challenging algorithms since they exctract the boundary elements belonging to the structure and integrate these elements into a coherent and consistent model of the structure.
In this section, two dimensional and three dimensional deformable models will be focused. In 3-D imaging, both of these approaches can be used. Ballons can themselves represent the surface structure of the 3-D volume while snakes represent the contours in each 2-D slice of 3-D volume and those contours can be stacked to form the surface.
4.1. Snake Model
A snake is an energy minimizing curve guided by external constraint forces and influenced by image forces that pull it toward the features such as lines and edges. The internal smoothing forces lock the snakes onto nearby edges. Initially, the snake is located near the image contour of interest and is attracted towards the target contour by the forces depending on the intensity gradient.
The idea behind the deformable contours is to associate an energy functional to possible contour shapes and then minimizing the functional for the detection of the contour. The energy functional can be represented as a sum of internal and external energies. Internal energies encourage continuity and smoothness of the contour. External energies on the other hand, influence the global structure of the contour attracting it to the defined features on the image. A demo of how the active contours converge to the real contours of the image is presented here.
Below, the mathematical formulation of the energy function is derived. Here
the integral is taken along the contour c, and and he energy terms are functions
of c or the derivatives of c. The parameters a,
b,
g
![]()
![]()

![]()
Different external and internal forces have been proposed to the initial description of the snake model. Splines have been fitted for smoothing constraints or other external forces such as interactive forces, or feature distance forces have been applied. Active contour model have been used for for segmentation of contours in 2-D slices of 3-D volumes[20]. The resulting sequence of 2-D contours can then be registered to form a continuous 3-D surface model.
4.2. Balloon Model
This approach is a 3-D extension of the snake model. This method also includes the internal and external forces that make the deformable model (balloon) converge to the boundaries of the volume. The internal forces act as smoothing terms while the external terms satisfy the global accuracy of the deformable model.
The balloon is represented as a vector valued parametric function, x (u,v) = [ x(u,v) , y(u,v), z(u,v)]T . where vector x denotes the positions of the material points. The deformation energy for the balloon model is given as follows :

where weighting functions a control the tensions in u and v directions, while b control the corresponding bending rigidities. A general approach to fit the deformable models to data is making the model time varying by adding a time variable to the formulation. The dynamic behavior of the balloon model during the convergence period is governed by the second order partial differential equations of Lagrangian dynamics formulation:
![]()
where µ represents the mass density. Here f represents the external forces that attract the model towards the features of the volume. They can be chosen as the image itself, or the edges in the image or some correlation window (especially for tracking purposes).
Next step in the deformable balloon model description is to discretisize the model. For this purpose Cohen used the finite element method, while Terzopoulos made a matrix representation of discrete equations of motion. Finite difference solutions approximate the continuous function x as a set of discrete nodes in space. To apply the finite element method, the continuous data is represented by a mesh. Different mesh representations have been used for the solution of the problem. The idea is to apply the internal and external forces to each node of the triangles representing the mesh and to further move the nodes of the triangles until the energy of the nodes are minimized.
In [26], deformable balloons have been used with different weighting functions of internal and external forces. The external forces were divided into feature forces and the data forces. Every feature of the image was linked to every point of the surface, so that the model converged to the surface itself regardless of the initial position of the model itself. For the data forces however, only the closest data point was taken into account.
The balloon model was shown to be robust in applications of medical image segmentation, face representation and tracking of three dimensional data.
5. Representation and Matching
The next step in the recognition process is the representation of the segmented structures. In this section, we will mainly focus on two main boundary representation schemes : Fourier Descriptors and Polynomial Fitting. Both of these methods represent the boundary with a vector, and the matching operation can be implemented by an Euclidean distance measure.
5.1. Elliptic Fourier Descriptors
Elliptic fourier Descriptors have been shown to be extremely powerful in the representation of closed boundaries by only a few coefficients. Any closed contour in 2D can be represented by two periodic functions of t, where t varies from 0 to 2pi, x(t) and y(t) which can in turn be expressed by their Fourier expansions in matrix form as [9]:
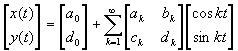
where :
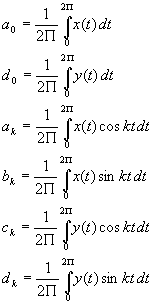
A shape is thus decomposed into a sum of rotating phasors, each defining an ellipse and rotating with a speed proportional to their harmonic number. The a, b, c, and d parameters uniquely define the ellipse. The invariants involving these parameters can be found as follows:
![]()
A demo showing the realization of contours by an addition of ellipse is shown here. To match two objects which were represented by elliptic Fourier Descriptors each, we can simply look at the distances of invariants of Fourier coefficients. SSD matching can be directly applied to the Fourier invariants.
5.2. Polynomial Based Representation
An algebraic curve is defined as the zero set of a polynomial of two variables. Similarly 3D surface boundary of an object can be represented by a zero set of a polynomial in three variables. The problem involved in polynomial based representation, is to find the best polynomial which can be fitted to a set of points from the segmentation phase.
Algebraic curves or surfaces are extremely powerful for shape recognition. The advantages of polynomial based representation over Fourier Descriptors is its applicability to open or non ordered data that may contain gaps. In this section, the methods for polynomail fitting to a set of points will be covered[25].
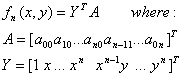
If we evaluate the above equation in each point of the data set and minimize the residue of the fitted polynomial, the formulation becomes:

The solution to the coefficients of the polynomial is given by the unit eigenvector of S associated with the smallest eigen value of S. This solution is invariant to affine transformations.
6. Discussion and Conclusion
Computation of accurate geometric shape from 3D medical images is a recent problem, and the solution of this problem would achieve a high improvement in the diagnosis and treatment of the diseases. Geometric shapes of the medical objects are highly complex and nonregular. Deformable models have been proposed to deform into a broad variability of anatomical structures. In this section, a general discussion of the applicable techniques to the problem of medical image segmentation, representation and matching will be given.
Due to the complex shapes of medical images, the local and the global views of the image should be considered. For our problem of 3D image segmentation, deformable models promise to be effective since they use a hybrid of the local and global properties of the images. The proper selection of the internal forces (smoothing forces) and external forces (feature and data forces) and also the weight functions is required for the robustness of the algorithm.
For the 3-D balloon fitting, the mathematical formulation that was generated by T.Mcinerney and D. Terzopoulos can be used [6]. In this paper the problem is solved using the analogy to the problem of Lagrange motion and force formulations. The gradient of the image is the usual and good choice for the external forces[7]. Thresholding following the gradient operation would give an estimate of the edges in the volume. In figure 2, the edges of some CT images are shown. The distance to the closest edge point can be used as a measure of external forces that deform the model globally to the shape.
After the deformable model fits the shape, the next step is to represent the deformable with some parameters. Polynomial fitting and Elliptic Fourier Descriptors methods can be used for this purpose. Both use the mesh structure of the converged deformable model. Using one of the techniques described in section 5, the shape can be represented by only a few coefficients. Elliptic Fourier Descriptors (EFD) seems to be the better choice due to the smooth shapes of the medical objects. The formulation of 2-D Elliptic Fourier Descriptors can easily be extended to higher degrees[27]. A measure for the amount a particular harmonic effects the reconstructed shape, can be used for the elimination of unnecessary harmonics and to keep the least necessary amount of data [28]. In [28], SSD measure was used to solve the matching problem between the library model's EFD coefficients, and tested model EFD coefficients.
Another improvement in the application of deformable models to shape classification is model based deformation. Initialization is one of the problems in deformable model theory. In theory, after the model library shapes are deformed, the deformed models can be used as the initial deformable shapes in the process of testing new polyp like structures. By using the model based initialization, additional measures such as the number of iterations or the effort in the iterations can be used.
Global Viewed, non deformable approaches can also be used for the representation and the matching problem. Fourier transformation of the surface itself is a way to compare the shape structures. The advantage of this approach is the reduction of the segmentation step. Even the volume itself can be used for the Fourier transformation and Fourier transformation properties can be used to find the Fourier Transformation of the gradient of the image as shown in section 3.2.
Another approach that can be used is the 4-dimensional application of Elliptic Fourier Descriptors, where x, y, z and the intensity value form the 4 dimensions. The advantage of this approach is that it uses both the shape and the density information about the polyps.
The discussed methods promise to be applicable for the solution of the segmentation and shape classification problem for the medical CT images. The results of the proposed methods will appear in my homepage soon.
Figure 2 - Edges on the Colon
Bibliography & References:
1 R. Jain,
R. Kasturi, B.G.Schunck, Machine Vision, Mcgraw Hill Interational Editions,1995.
2
A.Singh, D. Goldgof, D. Terzopoulos,Deformable Models in Medical Image
Analysis, IEEE Computer Society Press,
1998.
3 E. Trucco, A. Verri, Introductory Techniques
for 3-D Computer Vision, Prentice Hall, 1998.
4
T.Mcinerney and D.
Terzopoulos, Deformable models in Medical Image Analysis, Medicel image
Analysis, Volume 1, Issue 2:1996/7.
5 B.
Kass, A. Witkin, and D. Terzopoulos. Snakes: Active Contour models.
international Journal of Computer Vision 1(4) : 321-331. 1987.
6 T.Mcinerney and D. Terzopoulos, A Dynamic
finite Element Surface model for Segmentation and Tracking in Multidimensional
medical images with Application to Cardiac 4D Image Analysis, Journal of
Computerized Medical Imaging and Graphics, 1994.
7 G. Szekely, A. kelemen, Ch. Brechbuhler, and
G. Gerig, Segmentation of 2-D and 3-D Objects from MRI volume data using
constrained elastic deformations of flexible Fourier Surface Models, Medical image
Analysis, 1996
8 L.H. Staib, J.S. Duncan,
Deformable Fourier models for surface Finding in 3D images. Second Conference on
Visualization in Biomedical Computing, 1992, volume
1808,90-104.
9 L.H. Staib, J.S. Duncan,
Left Ventricular Analysis from Cardiac Images using Deformable models, Computers
in Cardiology, 1988. Proceedings. , 1989 , Page(s): 427 -430
10
R. Duda and P. Hart, Pattern
Classification and Scene Analysis, Wiley-Interscience Publication, 1973.
11
R.C. Gonzales, R.E.Woods, Digital Image Processing, Addison Wesley,
1993
12 L.H. Staib, J.S. Duncan,
Parametrically Deformable Contour Models, Computer Vision and Pattern
Recognition, 1989. Proceedings CVPR '89., IEEE Computer Society Conference on ,
1989 , Page(s): 98 -103.
13 S.B.
Gokturk, I. Bozma, L.Akarun, Automated Inspection of PCBs Using a Novel
Approach, Nonlinear Signal and Image Processing'99.
14
Bracewell, Fourier Transformation and its applications, McGrawHill
Series, Second Edition, 1986.
15
L. Garcia, Probability and Random Processes for Electrical Engineers,
Addison Wesley, 1994.
16 A.G.Mamistvalov,
n-dimensional moment invariants and conceptual mathematical theory of
recognition n-dimensional solids,Pattern Recognition, 1990. Proceedings., 10th
International Conference on, Volume: i , 1990 , Page(s): 288 -290 vol.1
17
R.J.Cambell, P.J.Flynn, Eigen shapes for 3D Object Recognition in
Range Data
18
J.Ben-Arie, D.Nandy, A Volumetric/Iconic Frequency Domain
Representation for Objects with application for Pose Invariant Face Recognition,
IEEE Transactions on Pattern Analysis and Machine Intelligence, Vol. 20, No 5,
May 1998.
19
A. lundervold, N. Duta, T. Taxt, A.K.Jain, Model guided Segmentation
of Corpus Collasum in MR Images, IEEE Conference on Computer vision and Pattern
Recognition'99, Vol 1, 231 - 237.
20
C.W.Chen, L.W.Chang, J.R.Ho, Reconstruction of 3D Medical Images: A
nonlinear interpolation technique for reconstruction of 3D medical images.
Computer Vision, Graphics and Image Processing 53(4):382-391.
21
R.Vogt, Precise Extraction of Bones from CT Scans, Advances in
Mathematical Morphology, Volume 2.
22
J.Serra, Image Analysis and Mathematical Morphology, Academic Press,
New York, N.Y., 1982.
23
W.J.Niessen, K.L.Vincken, J. Weickert, M.A.Viergever, Three
Dimensional MR Brain Segmentation, International Conference on Computer Vision'
98. Sixth International Conference on , 1998 , Page(s): 53 -58.
24
C.Westin, A. Bhalerao, H. Knutson, R. Kikinis, Using Local 3D
Structure for Segmentation of Bone from Computer Tomography Images, Computer
Vision and Pattern Recognition, 1997. Proceedings., 1997 IEEE Computer Society
Conference on , 1997 , Page(s): 794 -800
25
T.Tasdizen, J.Tarel, D.B.Cooper, Algebraic Curves that Work Better,
IEEE Conference on Computer Vision and Pattern Recognition'99, IEEE Computer
Society Conference on , 1999 , Volume 2,Page(s): 794 -800
26
H.Delingette, M.Hebert, K.Ikeuchi, Shape Representation and Image
Segmentation Using Deformable Surfaces, Computer Vision and Pattern Recognition,
1991. Proceedings CVPR '91., IEEE Computer Society Conference on , 1991 , Page(s):
467 -472.
27
M. Wu,H. Sheu, Representation of 3-D Surfaces by two variable Fourier
Descriptors,Pattern Analysis and Machine Intelligence, IEEE Transactions on,
Volume: 20 8 , Aug. 1998 , Page(s): 858 -863.
28
S.B. Gokturk, H.Yalcin, H.I. Bozma, Remote Control Device
Classification using Elliptic Fourier Descriptors and Local Curvature
properties, Project Report For BEKO, Turkey.