STOCHASTIC ROADMAP SIMULATION SOFTWARE
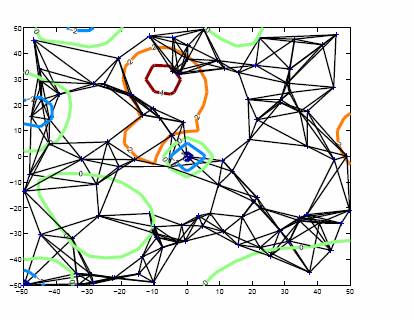
Figure 1: A sample roadmap in a
hypothetical 2-D energy landscape. The contours correspond to varying energy
levels, and thick black lines correspond to edges between nodes.
Many essential biological processes --e.g., protein folding and ligand-protein binding-- depend on the ability of molecules to move and adopt different shapes over time under the influence of potential energy fields. Computational techniques play an increasing role in the analysis and understanding of such motion. In particular, Monte Carlo Simulation and molecular dynamics methods are classic techniques for simulating molecular motion. But they have two major drawbacks:
v
They compute individual pathways, one at a time;
however, many interesting properties of molecular motion, in particular, the ensemble
properties, are best characterized statistically over many pathways. For
instance, the new view of protein folding hypothesizes that proteins fold in a
multi-dimensional energy funnel by following a myriad of pathways, all leading
to the same native structure. So we need efficient algorithms that can quickly
explore a large number of pathways.
v
A typical molecular energy function may contain many
local minima, and classic simulation techniques waste considerable computation
time trying to escape from these minima. They easily get trapped in local minima,
repeatedly sampling many similar conformations without obtaining much new
information. Their high computational cost prevents them from being used to
analyze many pathways.
We have developed Stochastic Roadmap Simulation (SRS) as a novel computational framework to overcome both of these drawbacks. In SRS, we build a network, called stochastic conformational roadmap, or just roadmap for short (see above figure for an illustration). Such a roadmap is a directed graph, whose nodes are randomly sampled molecular conformations. Each edge between two nodes in the roadmap carries a weight, which estimates the probability for the molecule to transition. A path between any two nodes in the roadmap corresponds to a potential motion pathway of the molecule. A roadmap thus compactly encodes a huge number of pathways. The edge probabilities determine the likelihood that the molecule may follow these pathways. SRS does not trace any specific pathway on the roadmap, and thus circumvents the local minima problem encountered with the classic simulation techniques.
Below you will find two applications of SRS to a 2-D hypothetical synthetic landscape and a vector based protein representation, along with some sample files.
Please send me an e-mail if you download the software, or have any questions. Thanks!
1. SRS for synthetic energy landscape (770 KB) : software to construct a synthetic energy landscape, and to compute the probability of folding on it. Along with matlab scripts for visualization, comparison with Monte Carlo simulation results, etc.
The README is separately also available.
2. SRS for vectorbased protein representation (2700 KB): same as above, except this is for vectorbased protein representation.
The README is also separately available.
3. Code for generating the input files for the vectorbased protein representation, for any protein: (450 KB)
The corresponding README.
4. SRS for ligand-protein docking (21.5 MB)
The corresponding README.
Reference:
1- Stochastic Roadmap Simulation: An efficient Representation and Algorithm for Analyzing Molecular Motion
M.S. Apaydin, D.L. Brutlag, C. Guestrin, D. Hsu, J.C. Latombe and C. Varma, Journal of Computational Biology, (10) 257-281, 2003.